
AbvarDB is a database designed to support researchers and clinicians in analyzing antibiotic resistance and susceptibility profiles in bacterial strains. The database is curated from the BVBRC database and integrates SNP data, allowing for detailed genetic analysis.
Utilizing tools like SwiftG and PolyPhen with VCF files, we analyze the functional impact of mutations within the Acinetobacter baumannii genome. The SIFT tool assesses amino acid substitutions to determine if they are deleterious, enhancing the accuracy of mutation analysis in AbvarDB.
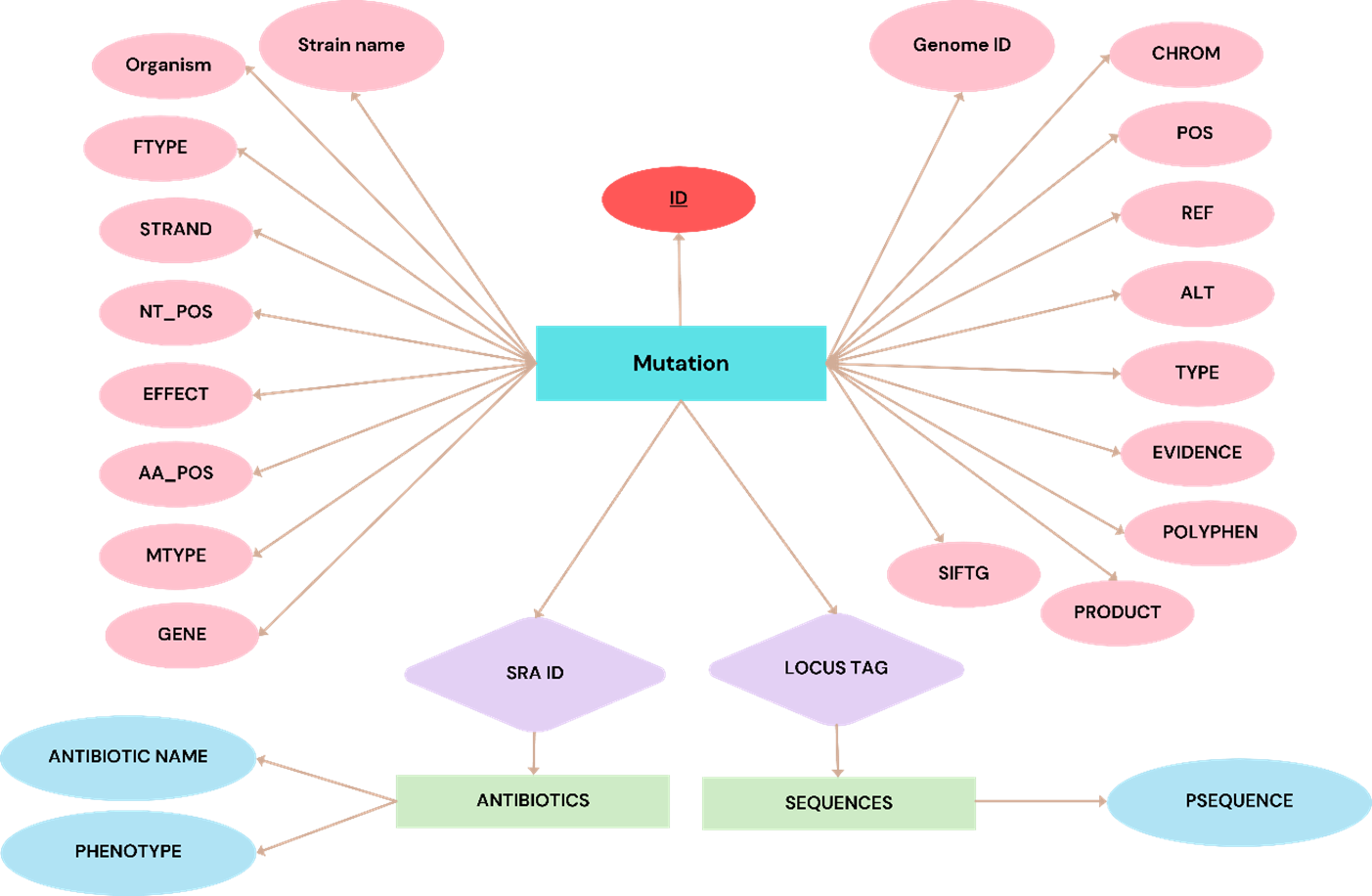
The AbvarDB database consists of three interconnected tables: Variant, Antibiotics, and Sequences. Each table contains specific information related to genetic variations, antibiotic resistance, and genetic sequences. The structure ensures robust data integrity and efficient data retrieval.
AbvarDB allows users to explore antibiotic resistance and susceptibility through two primary data categories: resistant and susceptible strains. The system is designed to be intuitive, offering multiple search approaches and visualization tools for easier data interpretation.
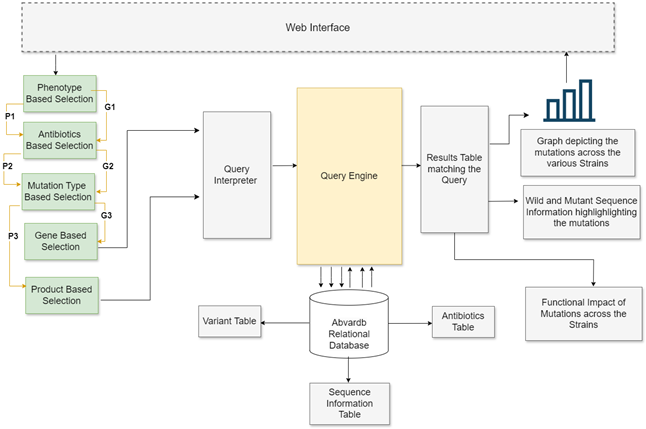
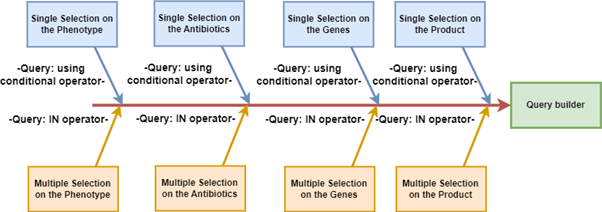
AbvarDB offers two main approaches for search: by phenotype (resistant or susceptible) and by antibiotics. Users can select mutation types, genes, or products, and the database generates comprehensive tables detailing information such as strain, gene, product, mutation, and the functional impact of mutations.
The system supports both single and multiple selections, using conditional operators for single selections and IN operators for multiple selections. This flexibility allows for tailored searches and efficient data retrieval.
AbvarDB employs Google Charts to visually represent mutations and their count across various strains. Users can click on a specific mutation to view the wild-type sequence alongside the mutant sequence, with color-coded annotations indicating deleterious and tolerant positions. The system also identifies strains containing a given mutation as resistant or susceptible, providing a bar graph displaying the count of mutations across strains.